Recently, my colleagues from Chinese Academy of Sciences published a paper about pumpkin TBBPA metabolites. I am one of the co-authors since I wrote the code to screen bronimated compounds. Later I wrote a formal function findohc in enviGCMS package to screen those compounds. The code design is slightly different with the original paper while it works fine. However, today my topic will not be about screen metabolites by mass defect clusters. I want to show you how to build a metabolites network by known PMDs.
The original question is simple and straightforward: it’s much easier to find metabolites for parent compounds while it’s not easy to find the metabolites of metabolites. In the recently published paper, my colleague showed the following figure:
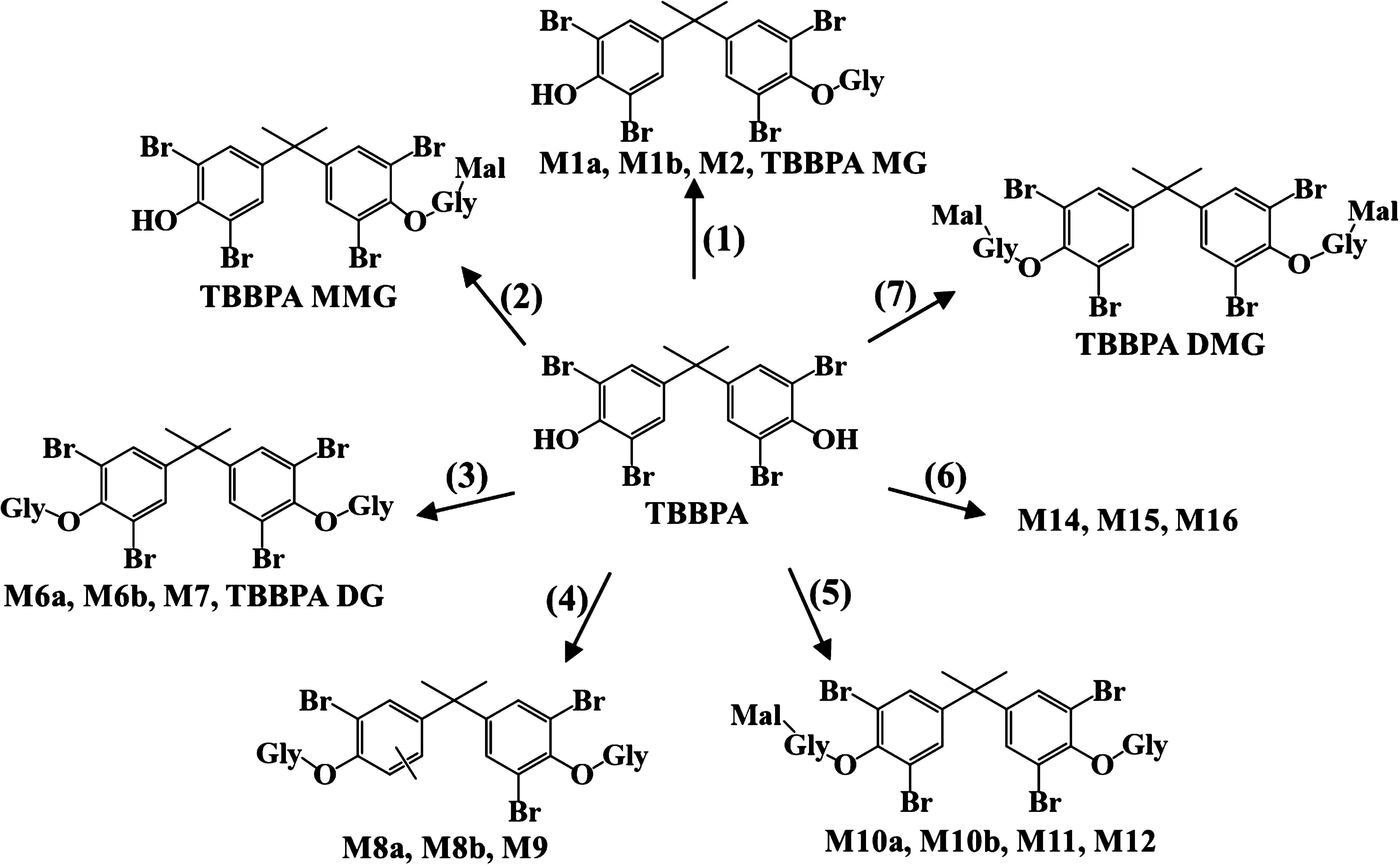
I agree with this plot while it obviously that some metabolites are from other metabolites instead of parent compounds. For example, TBBPA MG could be the parent compound of TBBPA MMG. It’s biologically meaningful to have those connected reactions. Then I suddenly realized my PMD analysis should also worked.
For pollution metabolism, Phase I and Phase II reactions are always there since both drug and pollution are treated equally as xenobiotic for living system. In this case, we actually know the corresponding PMDs. Here I selected five PMDs: “+Br/-H” for debromination process, “+6C10H5O” for Glycosylation, “+3C2H3O” for Malonylation, “+C2H” for Methylation and “+O” for Hydroxylation. I put atom number in front of atom character to make a difference with chemical formula. By writing a function called getchain to recursive search ions with those PMDs relationships in pmd package and filtered by pair-wise correlation coefficients larger than 0.6, I get this figure:
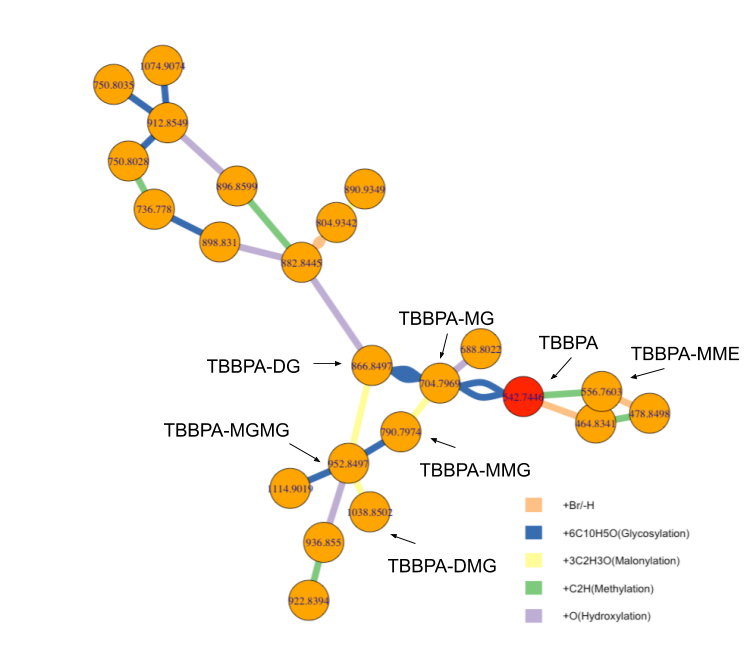
Here we could see metabolites network for TBBPA. It actually found lots of intermediate compounds from the raw data. Assuming those five reactions could stand for the major metabolism pathway in pumpkin, they could already produce a network structure. We could find TBBPA is not the center of the metabolism and Glycosylation build a bridge for more metabolites. The red dash line parts included some compounds we missed in the published paper. It’s surprising to find some Phase II reaction happened again and again. Since this work is already published, I didn’t expect another publication for this work. However, I believe this script would be useful for community. In this case, I included peaks list data from this study in enviGCMS package. Here is the code to reproduce the network analysis for TBBPA metabolites and please cite the papers for pmd and enviGCMS package, as well as the TBBPA study if you use those functions:
remotes::install_github('yufree/enviGCMS')
remotes::install_github('yufree/pmd')
install.packages(c('igraph','RColorBrewer'))
library(enviGCMS)
library(pmd)
# load the data
data(TBBPA)
# extract brominated compounds
x <- findohc(TBBPA)
brcomp <- getfilter(x,rowindex = x$mz %in% x$ohc$mz)
pmd <- list(data = brcomp$data,mz = brcomp$mz, rt = brcomp$rt, group = brcomp$group)
# build the network
df <- getchain(pmd,diff = c(77.91,162.05,86,14.02,15.99),542.7446)
df$sdac$ms1 <- round(df$sdac$ms1,4)
df$sdac$ms2 <- round(df$sdac$ms2,4)
library(igraph)
library(RColorBrewer)
pal <- brewer.pal(5,"Accent")
# only use pairs with correlation coefficience larger than 0.6
net <- graph_from_data_frame(df$sdac[abs(df$sdac$cor)>0.6,],directed = F)
# show the network
plot(net,vertex.size =15,edge.color = pal[as.numeric(as.factor(E(net)$diff2))],edge.width = 10)
legend("topright",bty = "n",
legend=c('+Br/-H','+6C10H5O(Glycosylation)','+3C2H3O(Malonylation)','+C2H(Methylation)','+O(Hydroxylation)'),
fill=unique(pal[as.numeric(as.factor(E(net)$diff2))]), border=NA)
Besides, I will attend American Chemical Society Fall 2019 National Meeting & Expo in San Diego, CA next month and give a oral presentation about structure/reaction directed analysis for environmental studies. If you will be there and want to do some stupid cool things for untarget analysis, feel free to contact me.